Channing House Data¶
This is the channing dataset in the R package
boot. From
the package description:
Channing House is a retirement centre in Palo Alto, California. These data were collected between the opening of the house in 1964 until July 1, 1975. In that time 97 men and 365 women passed through the centre. For each of these, their age on entry and also on leaving or death was recorded. A large number of the observations were censored mainly due to the resident being alive on July 1, 1975 when the data was collected. Over the time of the study 130 women and 46 men died at Channing House. Differences between the survival of the sexes, taking age into account, was one of the primary concerns of this study.”
These data feature left truncation because residents entered Channing House at different ages, and their lifetimes were not observed before entry. This is a biased sampling problem since there are no observations on individuals who died before potentially entering Channing House.
In [1]:
import matplotlib.pyplot as plt
import seaborn as sns
sns.set(style="darkgrid", palette="colorblind", color_codes=True)
from survive import datasets
from survive import SurvivalData
from survive import KaplanMeier, NelsonAalen
Loading the Dataset¶
The channing() function in the survive.datasets module loads a
pandas.DataFrame containing the Channing House data. The columns of
this DataFrame are
sex- Sex of each resident (male or female).entry- The resident’s age (in months) on entry to the centre.exit- The age (in months) of the resident on death, leaving the centre or July 1, 1975 whichever event occurred first.time- The length of time (in months) that the resident spent at Channing House (this isexit - entry).status- Right-censoring indicator. 1 indicates that the resident died at Channing House, 0 indicates that they left the house prior to July 1, 1975 or that they were still alive and living in the centre at that date.
In [2]:
channing = datasets.channing()
channing.head()
Out[2]:
| sex | entry | exit | time | status | |
|---|---|---|---|---|---|
| resident | |||||
| 0 | male | 782 | 909 | 127 | 1 |
| 1 | male | 1020 | 1128 | 108 | 1 |
| 2 | male | 856 | 969 | 113 | 1 |
| 3 | male | 915 | 957 | 42 | 1 |
| 4 | male | 863 | 983 | 120 | 1 |
Exploratory Data Analysis¶
We use the Channing House data to create a SurvivalData object.
In [3]:
surv = SurvivalData(time="exit", entry="entry", status="status", group="sex",
data=channing)
print(surv)
female
798+ 804 804+ 812+ 819+ 821+ 822 824+ 825+ 829+ 830 836+
840 845 848+ 848+ 854+ 857+ 860+ 861 861+ 868 870+ 870+
872+ 873 874+ 875+ 876+ 882+ 883 885 888+ 891+ 891+ 892+
893+ 895 895+ 897 897+ 898+ 899+ 901 904+ 905 905 905+
905+ 905+ 906+ 908 908 911 912+ 912+ 912+ 913+ 914+ 915
916+ 917+ 918+ 919 919+ 922+ 923 924+ 925+ 926 926+ 926+
926+ 927+ 927+ 927+ 928 928+ 929+ 930 930+ 931 932 932+
932+ 932+ 933+ 934 934+ 936 938+ 938+ 938+ 939+ 939+ 940
940+ 941 942+ 943+ 944 944 944+ 944+ 945+ 946+ 947+ 948
948+ 948+ 950+ 950+ 952+ 952+ 953+ 953+ 954 954+ 955+ 955+
955+ 957+ 957+ 958+ 958+ 959 959+ 959+ 960+ 961+ 961+ 962+
963 963+ 964+ 965+ 965+ 966 967+ 969 969 970 970+ 971+
971+ 973+ 973+ 975 975+ 975+ 976 976+ 976+ 976+ 977+ 977+
978 978+ 979+ 979+ 979+ 981+ 982 982 982+ 983 983+ 984+
985+ 985+ 985+ 985+ 986 986+ 987+ 988+ 988+ 989 989+ 989+
990 990 990 990 991 991+ 992 992+ 992+ 993+ 993+ 994
994 994+ 994+ 995 995 995+ 996 996 996 996+ 996+ 997+
998 998+ 999 1000 1000+ 1001 1001+ 1001+ 1003 1003+ 1004 1004+
1005 1005+ 1005+ 1006 1006 1006+ 1006+ 1006+ 1008+ 1008+ 1009+ 1010
1010+ 1010+ 1010+ 1011 1011+ 1011+ 1012 1012 1012+ 1012+ 1012+ 1013
1013+ 1014 1014+ 1014+ 1014+ 1015 1015+ 1015+ 1016+ 1017 1018 1018
1019 1019 1019+ 1019+ 1019+ 1019+ 1020 1020+ 1020+ 1021 1022+ 1023
1023 1023+ 1023+ 1024 1024+ 1027 1028+ 1029 1029+ 1030 1030+ 1031+
1031+ 1032+ 1033 1033+ 1034+ 1035+ 1036+ 1036+ 1037+ 1038+ 1040 1040
1040 1040 1041 1041 1041 1042+ 1042+ 1043 1043+ 1044 1044+ 1047
1047+ 1049+ 1050+ 1051+ 1053+ 1054+ 1054+ 1055+ 1056 1056 1057+ 1059+
1061+ 1062+ 1063 1063+ 1064 1065+ 1068 1068 1070 1071+ 1071+ 1072
1072+ 1073 1073+ 1074 1080+ 1083 1084 1085 1085 1085+ 1086 1088+
1089 1091+ 1093 1097 1102+ 1105+ 1105+ 1109+ 1114+ 1115 1119+ 1122
1131 1132 1134+ 1142 1147+ 1152 1172 1172+ 1186+ 1192 1200 1200
1207+
male
777 781 843+ 866+ 869 872 876 893 894 895+ 898 906+
907 909 911 911+ 914+ 927 932 936+ 940+ 943+ 943+ 945
945+ 948 951+ 956+ 957 957+ 959+ 960+ 966 966+ 969 970+
971 972+ 973+ 977+ 983 984+ 985 989 993 993 996+ 998
1001+ 1002+ 1005+ 1006+ 1009 1012 1012 1012+ 1013+ 1015+ 1016+ 1018+
1022 1023+ 1025 1027+ 1029 1031 1031+ 1031+ 1033 1036 1043 1043+
1044 1044+ 1045+ 1047+ 1053 1055 1058+ 1059 1060 1060+ 1064+ 1070+
1073+ 1080 1085 1093+ 1094 1094 1106+ 1107+ 1118+ 1128 1139 1153+
/Library/Frameworks/Python.framework/Versions/3.7/lib/python3.7/site-packages/survive-0.3-py3.7.egg/survive/survival_data.py:183: RuntimeWarning: Ignoring 5 observations where entry >= time.
RuntimeWarning)
The warning here is due to the fact that five of the observations in the data have entry times that are the same or later than the corresponding final times. These observations consequently cannot be used.
In [4]:
# The five observations responsible for the warning above
display(channing.loc[channing.exit <= channing.entry])
| sex | entry | exit | time | status | |
|---|---|---|---|---|---|
| resident | |||||
| 56 | male | 953 | 953 | 0 | 0 |
| 351 | female | 957 | 957 | 0 | 0 |
| 372 | female | 944 | 944 | 0 | 0 |
| 373 | female | 935 | 935 | 0 | 0 |
| 433 | female | 959 | 912 | 53 | 1 |
The last line above doesn’t even make sense since the entry time (959) is greater than the exit time (912). The resident’s duration is 53, which suggests that the entry time is likely supposed to be 859 (after all, 859+53=912). Nevertheless, we will ignore this observation.
In [5]:
display(surv.describe)
| total | events | censored | |
|---|---|---|---|
| group | |||
| female | 361 | 129 | 232 |
| male | 96 | 46 | 50 |
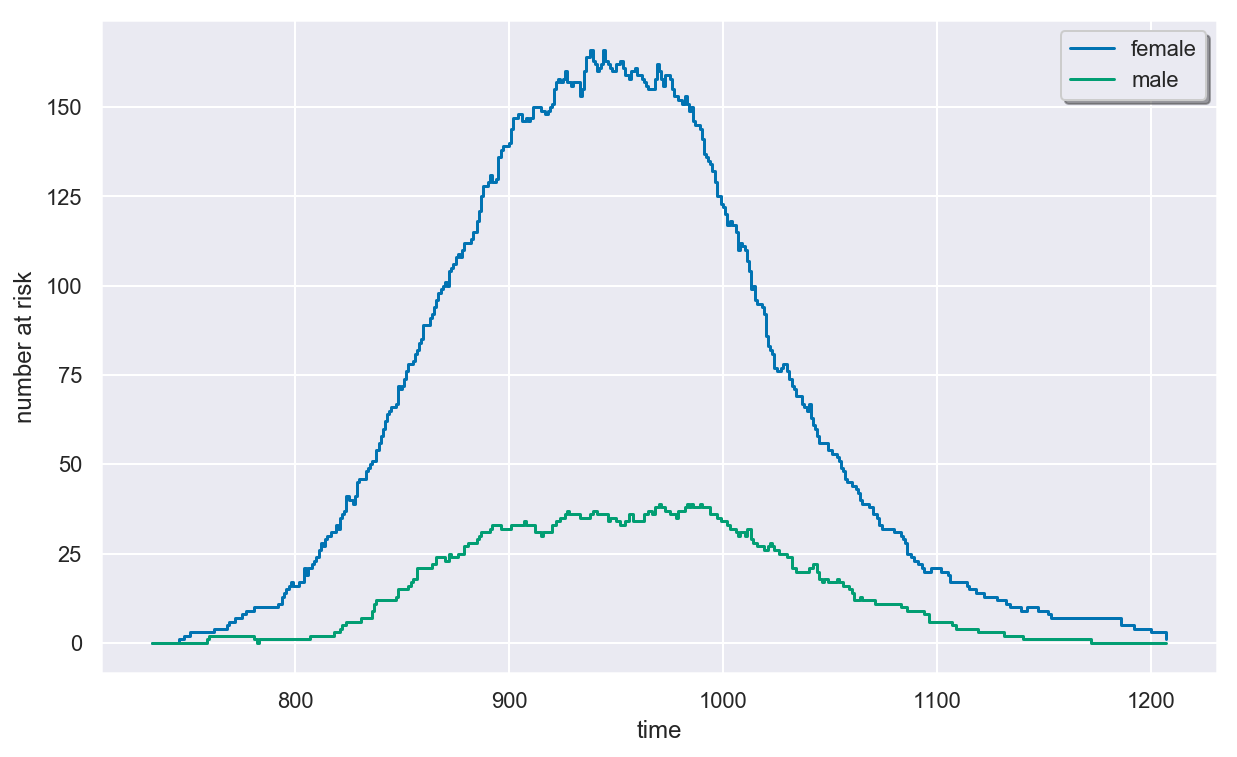
Plotting the At-Risk Process¶
Due to the left-truncation, the risk set size initially increases as residents enter Channing House and then decreases as residents die or are censored.
In [6]:
plt.figure(figsize=(10, 6))
colors = dict(female="b", male="g")
surv.plot_at_risk(colors=colors)
plt.show()
plt.close()
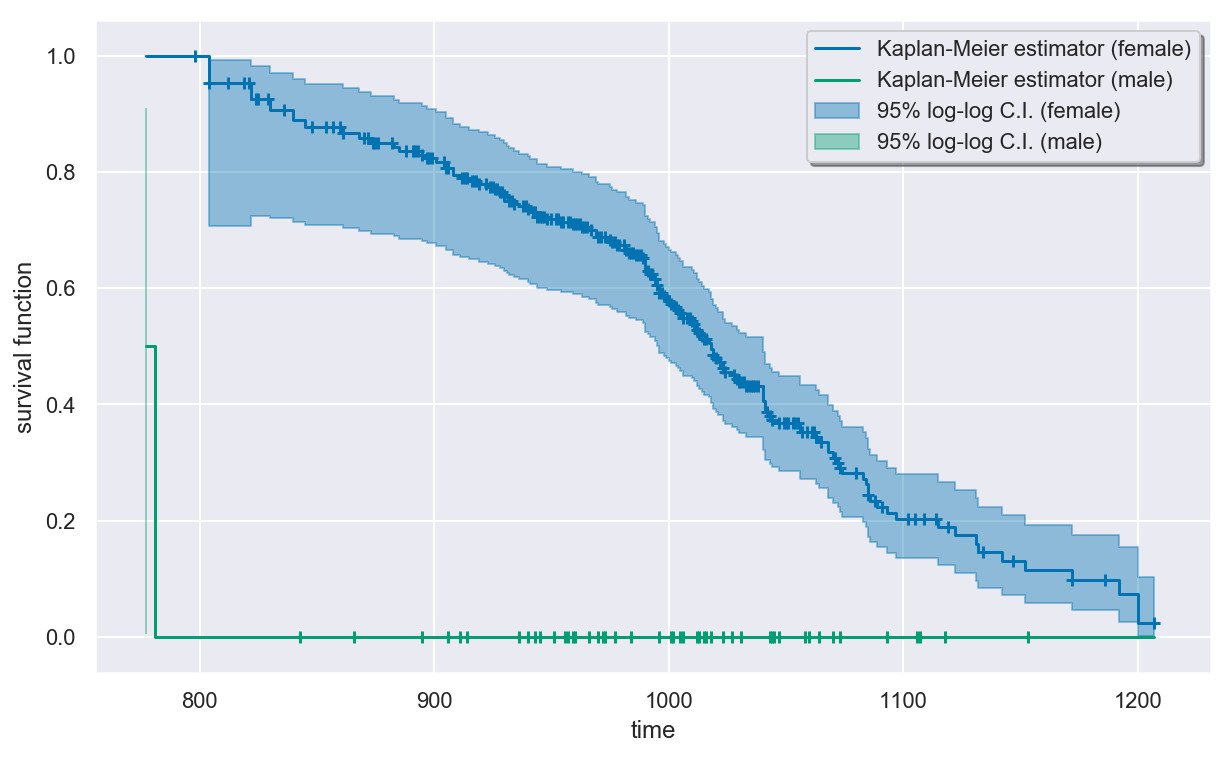
Estimating the survival function¶
The out-of-the-box Kaplan-Meier estimator does a poor job estimating the survival function for the Channing House data because risk set of the male residents is zero very early on before growing. This causes the survival function estimate to be zero for most of the observed times, which is clearly wrong. We will show the problem graphically in this case and then discuss how to fix it.
In [7]:
km = KaplanMeier().fit(surv)
plt.figure(figsize=(10, 6))
km.plot(colors=colors)
plt.show()
plt.close()
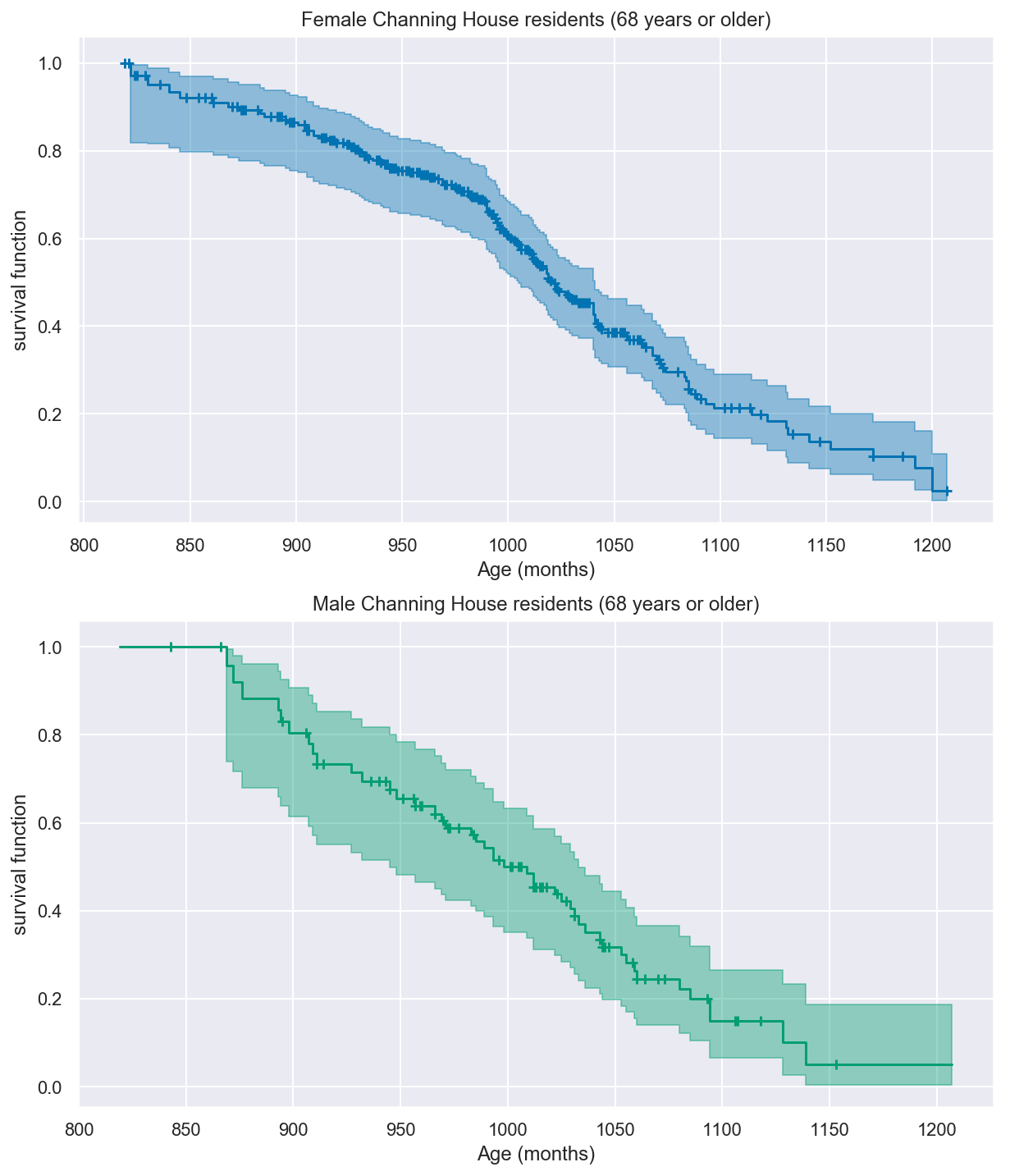
One way to address this issue is to condition on survival up to a later time, say 68 or 80 years (816 or 960 months).
In [8]:
km68 = KaplanMeier()
km68.fit(time="exit", entry="entry", status="status", group="sex",
data=channing, min_time=816, warn=False)
Out[8]:
KaplanMeier(conf_level=0.95, conf_type='log-log', n_boot=500,
random_state=None, tie_break='discrete', var_type='greenwood')
In [9]:
_, ax = plt.subplots(nrows=2, ncols=1, figsize=(10, 12))
km68.plot("female", color=colors["female"], ax=ax[0])
ax[0].set(title="Female Channing House residents (68 years or older)")
ax[0].set(xlabel="Age (months)")
km68.plot("male", color=colors["male"], ax=ax[1])
ax[1].set(title="Male Channing House residents (68 years or older)")
ax[1].set(xlabel="Age (months)")
plt.show()
plt.close()
In [10]:
km80 = KaplanMeier()
km80.fit(time="exit", entry="entry", status="status", group="sex",
data=channing, min_time=960)
Out[10]:
KaplanMeier(conf_level=0.95, conf_type='log-log', n_boot=500,
random_state=None, tie_break='discrete', var_type='greenwood')
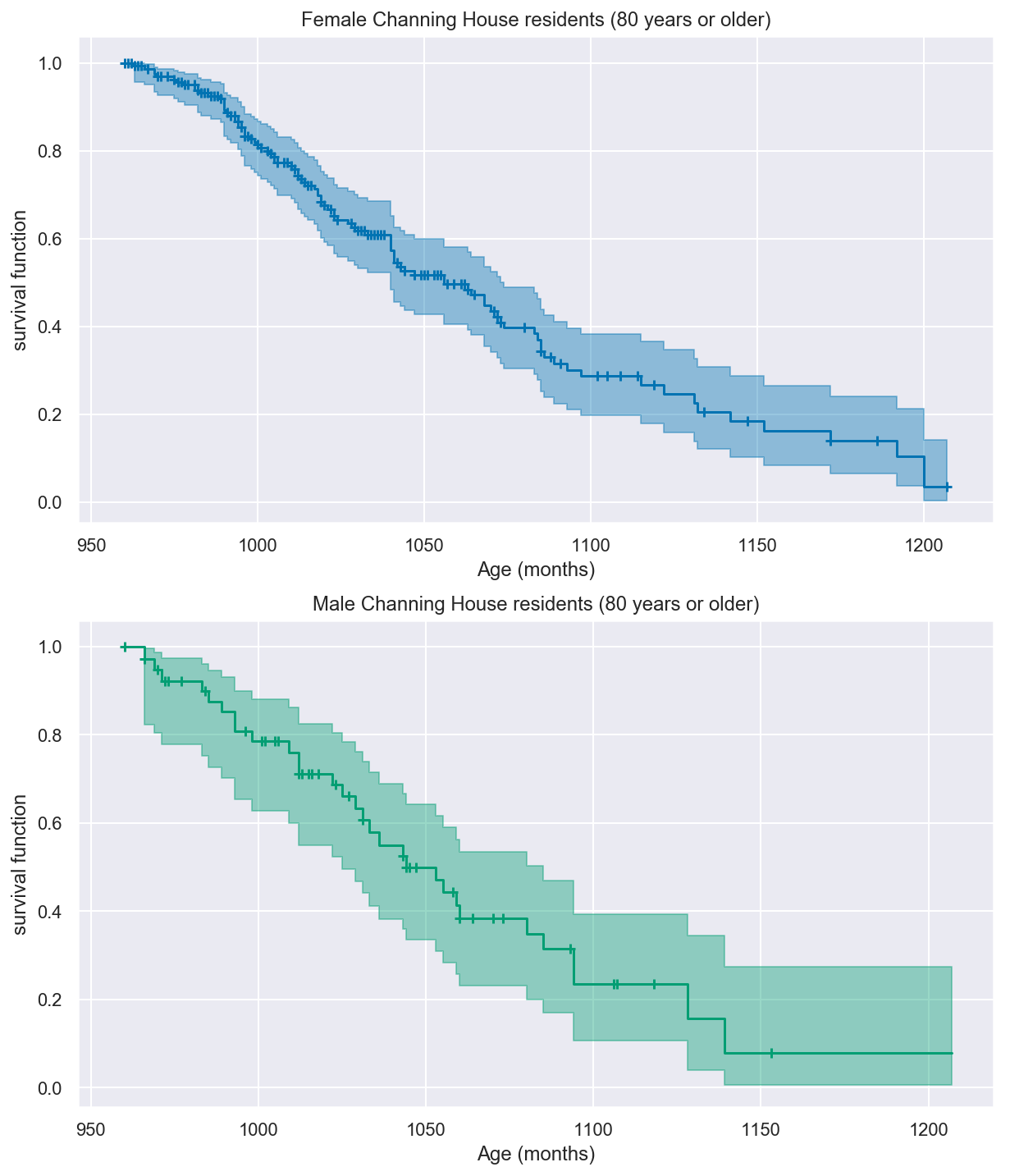
In [11]:
_, ax = plt.subplots(nrows=2, ncols=1, figsize=(10, 12))
km80.plot("female", color=colors["female"], ax=ax[0])
ax[0].set(title="Female Channing House residents (80 years or older)")
ax[0].set(xlabel="Age (months)")
km80.plot("male", color=colors["male"], ax=ax[1])
ax[1].set(title="Male Channing House residents (80 years or older)")
ax[1].set(xlabel="Age (months)")
plt.show()
plt.close()
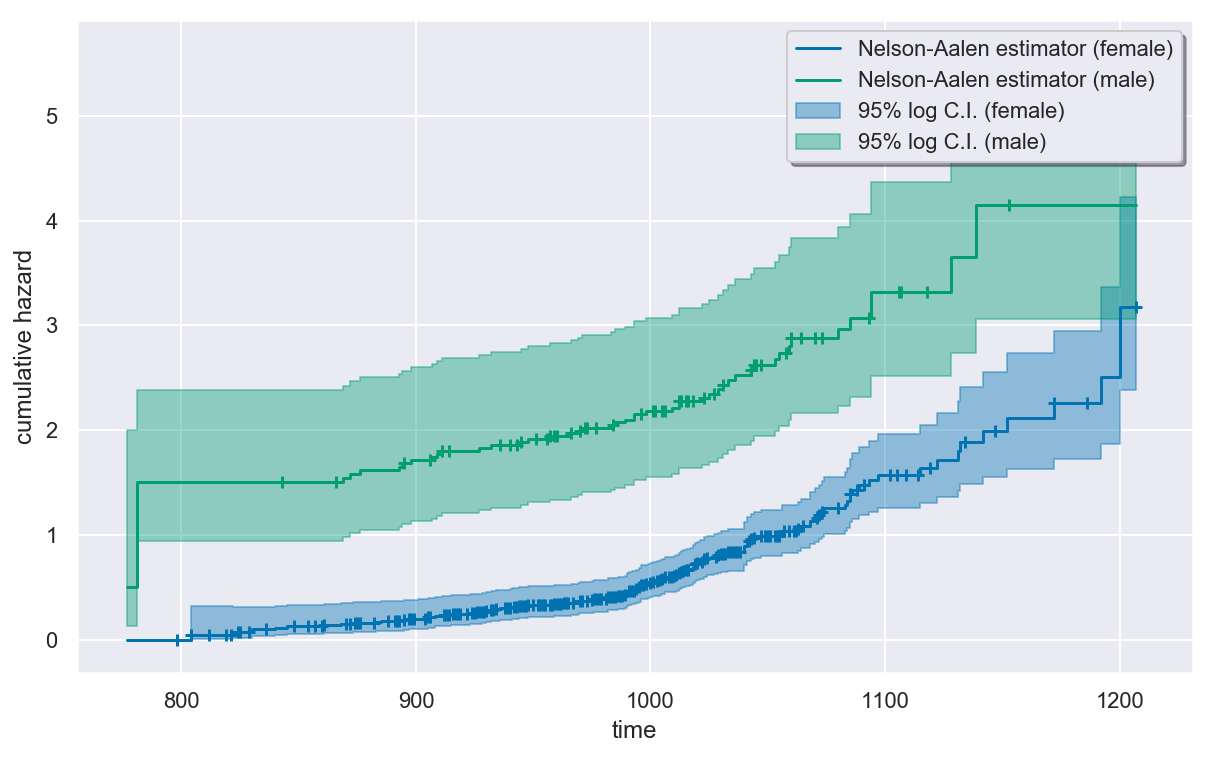
Estimating the cumulative hazard¶
In [12]:
na = NelsonAalen(var_type="greenwood")
na.fit(surv)
Out[12]:
NelsonAalen(conf_level=0.95, conf_type='log', tie_break='discrete',
var_type='greenwood')
In [13]:
plt.figure(figsize=(10, 6))
na.plot(colors=colors)
plt.show()
plt.close()